Epigenetics, misunderstood concept
Why mangling scientific terms is not OK
It’s hard for scientists to get funded and harder still to get good funding. Buzz words help scientists get that mullah. Epigenetics, a truly awe inspiring molecular process, has become a buzz word. Mangled, coopted and twisted, it can mean everything and anything.
This didn’t set well with a pioneer in the field called Mark Ptashne. He’s 80 years old and says things that make me like him, like
“The important point is to attend to how things actually work1
He’s not well known but should be. He’s the first scientist to show specific binding between protein and DNA. He also originated the ‘ball and stick’ model of transcription factors. Most notably, he elucidated the mysteries of the virus called bacteriophage lambda, an indispensable tool for biologists.
His colleagues have said that [bold = my emphasis] [1]
“Everythingwe know about gene transcription has come from...Ptashne”
Don’t get it twisted, this is no argument by authority. It is demonstrated performance (notice I didn’t cite his qualifications or awards). So what did Ptashne have to say about epigenetics? In his 2007 paper On the use of the word 'epigenetic' he said [2]:
“Over the past few years we have seen an odd change, or extension, in the use of the word ‘epigenetic’ when describing matters of gene regulation in eukaryotes. Although it may generally be that it is not worth arguing over definitions, this is true only insofar as the participants in the discussion know what each other means. I believe the altered use ofthe term carries baggage from the standard definition that can have misleading implications.”
This sort of definitional creep, like most crappy science, is a leap from correlation to causation. According to Ptashne
“Patterns of histone modification are often strongly correlated with patterns of inherited gene expression, and it has often been assumed that these modifications must cause them— hence the questionable use of the term ‘epigenetic’ to describe all such modifications.
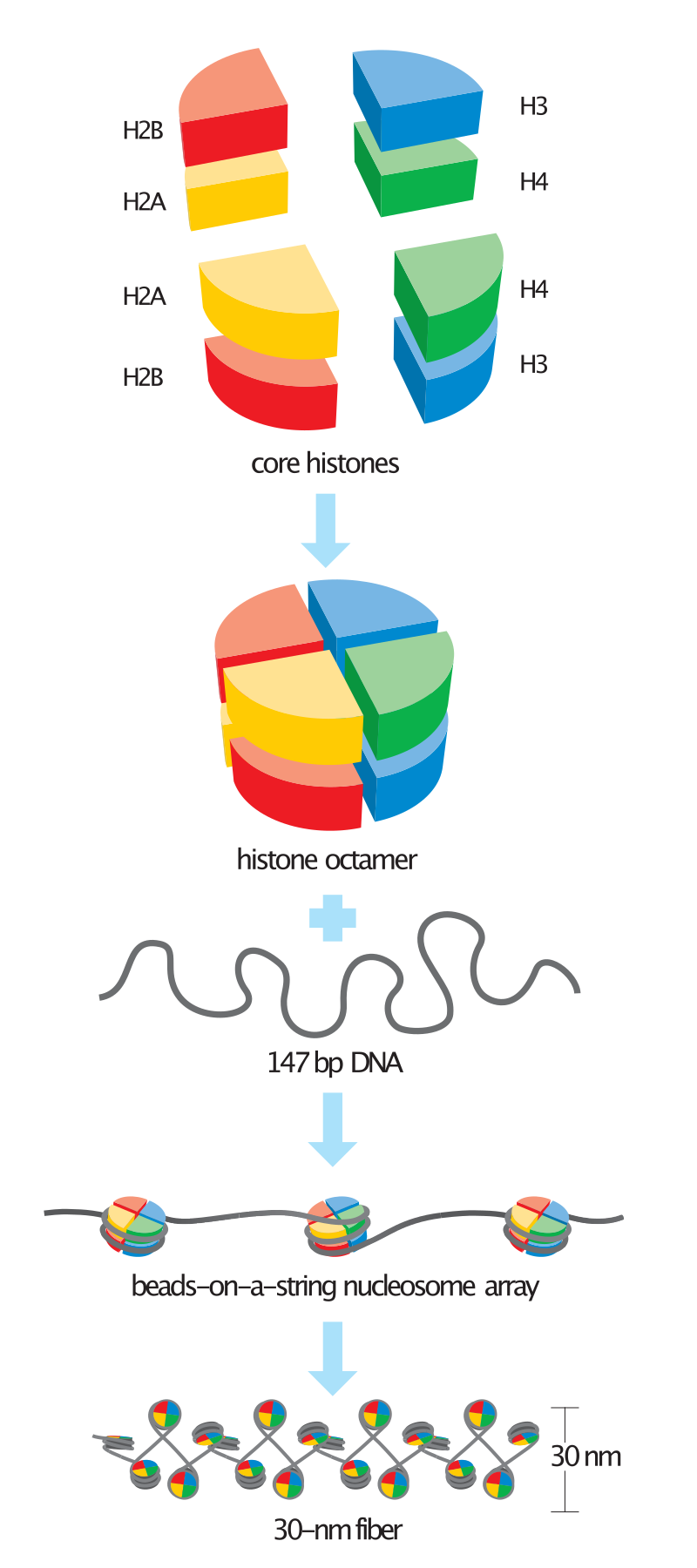
Histones are misunderstand in a second way. Before I tell you how though, keep in mind that they’re the stuff of nucleosomes around which DNA is wrapped in higher organisms like humans and garden worms (not bacteria).
Ptashne explains in his 2013 paper that the widely subscribed but incorrect view is that [3]
“chemical modifications to DNA (e.g. methylation) and to histones — the components of nucleosomes around which DNA is wrapped in higher organisms —drive gene regulation”
Nucleosomes cannot drive gene regulation because they don’t provide specificity. That means a unique sequence to be recognised by the enzymes performing the chemical modifications. Without it, they can’t distinguish individual nucleosomes or methyl groups. Point is, gene regulation requires specificity. In Ptashne’s words from 2007
“there areno known enzymes that recognize a specific modification on a nucleosomeand ensure that that modification will be transmitted to the next generation”
These enzymatically performed chemical modifications decay over time. Multigenerational maintenance requires memory or self-perpetuation (or self-propagating). Normal gene regulation doesn’t do that. And neither do nucleosome modification patterns or transcription factors. You’d have to remove molecular recruiters to see if your histone modifications self-perpetuating (and if you did you’d see they wouldn’t be). Ptashne is explicit
“there are no examples where these modifications have been shown to be self-propagating, and there are explicit examples wherethese modifications are shown not to be self-propagating”
However, he does mention how X inactivation/imprinting occurs in mammals across generations. Indeed, the black and orange calico cat coat pattern is unchanging. However we have no clue how they do that...
Furthermore,
“Just astranscription per se is not “remembered”without continual recruitment,so nucleosome modifications decay as enzymes remove them(the way phosphatases remove phosphates put in place on proteins by kinases), or asnucleosomes, which turn over rapidly compared with the duration of a cell cycle, are replaced”
So how does Ptashne define epigenetics?
specificity (e.g. gene apposed cis-regulatory sites)
self-perpetuating (e.g. positive feedback loop, double-negative loop or viral immunity transferred to sperm)
memory (i.e. the multi-generational persistence)
1 or even 2 of these mechanistic occurrences don’t count as epigenetic an phenomenon. That’s the mistake most papers with the word ‘epigenetic’ in them make. Like this one for example - I see you Nick Norwitz! It measured chromatin immunoprecipitation (i.e. nucleosome patterns) and RT-qPCR (i.e. gene expression) and then just assumed the changes in gene expressions were caused by the nucleosome patterns. The developmental phase of organism is also a case of mistaken epigenetic identity, as it happens in offspring without without being directly inscribed in genetic sequences.
It is thus when you get a sequence specific, self-perpetuating and multi-generational pattern that you have an epigenetic phenomenon.
Definition are better when they’re specific as can be. Without definitional rigor we run the risk of believing interventions to be multi-generational when they’re not. We could fail to deliver useful interventions to offspring. Conversely, we might fail to identify actual epigenetic phenomena, thus missing harms inherited by offspring. Forgive the tHinK oF The cHiLDreN! argument, but it’s true in this instance.
Epigenetic phenomena are real and observed in multiple organisms, like mice embryo and yeast, but few of them have been established with the rigour Mark Ptashne Brough to his experiments. He is our standard and we ignore it at our peril.
I’ll leave you with another gem from Mark Ptashne “a skeptic is someone who can imagine a better answer2. Lets stand on his shoulder and imagine a better answer.
Be well friends!
from ref 2
from ref 1



Fine post marred by epiphilological errors (typos).
Close the quote at the end.
Mark Ptashne Brough?
"You’d have to remove molecular recruiters to see if your histone modifications *are* self-perpetuating (and if you did you’d see they wouldn’t be).
More where they came from.
Fine post marred by epiphilological errors (typos).
Close the quote at the end.
Mark Ptashne Brough?
"You’d have to remove molecular recruiters to see if your histone modifications *are* self-perpetuating (and if you did you’d see they wouldn’t be).
More where they came from.